The Data Sets - Hippocampal CA3 Interneuron
Folder Contents:
This is 1 of the 6 data sets used for DIADEM.
This data set consists of 2 separate neurons, ‘Neuron 1’ and ‘Neuron 2’. ‘Neuron 1’ and ‘Neuron 2’consist of 5 and 4 image stacks, respectively, each containing dendritic and axonal subtrees from a different hippocampal CA3 interneuron. The material for each interneuron was sectioned into physical slices, each of which is captured by one of the image stacks. Axonal and dendritic arbors are divided into sections and thus are broken up by section.
During the DIADEM Grand Challenge Competition: ‘Neuron 1’ was used for the Training (Section 1) and Qualifier (Sections 2 though 5) Rounds and ‘Neuron 2’ was used for the Final Round.
Experimental Procedures:
data set owner: G Barrionuevo, Department of Neuroscience, University of Pittsburgh
Species: Rat
Strain: Sprague-Dawley
Nervous System Region: Hippocampus CA3
Fiber type: Dendrites and Axons
Labeling Method: Biocytin
Image Acquisition Method: Transmitted Light Brightfield
Tracing Method: Neurolucida (Williston, VT)
Objective Lens: 100x oil
See Calixto et al., 2008, J Physiol. Vol 586(11) for general information related to the data
Z distance between successive images within an image stack: 1.52 pixels
Download Instructions:
1. Download and extract 'Hippocampal CA3 Interneuron Part_1.rar'
through 'Hippocampal CA3 Interneuron Part_7.rar'. Note that these RAR files are independently compressed and must be extracted individually.
2. All RAR files contain the folder ‘Neuron 1’ or ‘Neuron 2’, each containing a portion of the data from these folders. Extracted RAR files should be merged to have the following folder hierarchy: Create the parent folder ‘Hippocampal CA3 Interneuron’; copy all data into the subfolders ‘Neuron 1’ and ‘Neuron 2’.
3. ‘Neuron 1’ ‘Image Stacks’ folder will contain 5 subfolders: ‘Section 1’ through ‘Section 5’. ‘Neuron 2’ ‘Image Stacks’ folder will contain 4 subfolders: ‘Section 1’ through ‘Section 4’.
4. Each ‘Section x’ folder contains one image stack, except for ‘Neuron 2’, where 'Section 2' contains 1 image stack called 'Full size' and one called '75% size'. 'Section 4 contains one image stack called 'Full size' and 1 called '90% size'. '75% size’ and '90% size' are explained in instruction 12 below.
5. All reconstructions will automatically align with their respective image stacks when loaded into software that uses pixel-based coordinates, such as ImageJ or Neuromantic. Use ‘Gold Standard Starting Coordinates' below to locate tree samples.
6. Image stacks may contain overlapping structures that are separate from the given tree sample and should not be traced.
7. The starting coordinates given below clearly mark the root of each tree sample to reconstruct within its given image stack section.
8. Special note for this data set: Some tree samples do not bifurcate; consisting of only a root and a single termination node.
9. Special note for this data set: All gold standard reconstructions of tree samples were manually reconstructed in 3D, but the Z coordinates in between bifurcation nodes, terminations, and root were smoothed (i.e. interpolated) prior to submission. Therefore, automated reconstructions should use linear connections between all branch segments (i.e. between bifurcations, and terminations, and root) in order to compare to the gold standard reconstructions when using the DIADEM metric.
10. Special note for this data set: External branches (i.e. branches that end in a termination) with path lengths of less than 23 pixels will be ignored when using the DIADEM metric. Thus, they can be traced or ignored without affecting the DIADEM metric score, provided that the traced branches remain less than 23 pixels in length.
11. Special note for this data set: Neuromantic v1.7.5 provides certain advantages for some of the other smaller data sets however may prove counter-productive for use with this data set due to its dynamic image loading method. Neuromantic v1.6.3, which was the latest version at the time of the DIADEM Training/Qualifier Rounds, uses a different loading method and may work better for some of the image stacks in this data set.
12. Special note for this data set: In organizing gold standard reconstructions for this data set for DIADEM, a systematic method was used to decide where structures ended when they leave a given section:
A. If a tree drifted out of focus as it left a section (i.e. lost focus near the top or bottom images), the trace is terminated at the last point in focus.
B. If a tree appears to drift out of focus in the middle of a section for some reason, the trace is terminated further down to where the structure is minimally visible.C. If a tree ends either continuously or abruptly but seems to reappear downstream in the same section, suggesting either a gap in labeling or that it left the section and came back, then:
i. If the gap is small enough that one can be certain that the tree continues, then the tree is traced as continuing.
ii. If the gap is too large to be certain that the branches connect, the currently traced tree is not connected to the reappearing branch.
iii. If the gap shows the branch continuing but out of focus to the point of not being able to distinguish potential arborizations, the trace is also terminated prior to the out of focus structures and not connected if it appears in focus downstream.
iv. If the gap shows the branch continuing and out of focus but visible enough to see proper connectivity, then the trace is continued through the gap, ultimately connecting the reappearing structure downstream.
Ultimately, these decisions are somewhat subjective, and one should utilize the provided gold standard reconstructions to determine the tracing style and methods used.
13. Special note for this data set: Section 2 and 4 of ‘Neuron 1’ contain both full and scaled image stacks. Full size image stacks are too large to load in Neuromantic v1.6.3 and can cause significant time lag when scrolling through images using Neuromantic v1.7.5. Image stacks and corresponding gold standard reconstructions scaled to 75% of full size for ‘Section 2’ and 90% for ‘Section 4’ are provided and will load with no problems in Neuromantic v1.6.3. Note that image stacks were scaled in X,Y only. Scaled down image stacks for ‘Neuron 1’ are not provided even though some of them may be too big to fully load in Neuromantic v1.6.3. ImageJ can be used to create scaled down image stacks, while Neuromantic can be used to scale down corresponding gold standard reconstructions.
14. Special note for this data set: Imaging artifacts appear in a small portion of images. Images affected for ‘Neuron 2’ include: ‘Section 1’- ‘004.tif’ through ‘006.tif’ and ‘Section 2’ (both full and 75% scale sizes)- ‘013.tif’ through ‘023.tif’. Artifacts may not affect algorithm performance, however, it is suggested that if you choose to remove these images from the stack and replace them that you find some way to preserve respective Z values within associated image stacks. ‘Neuron 1’ was not checked for similar artifacts.
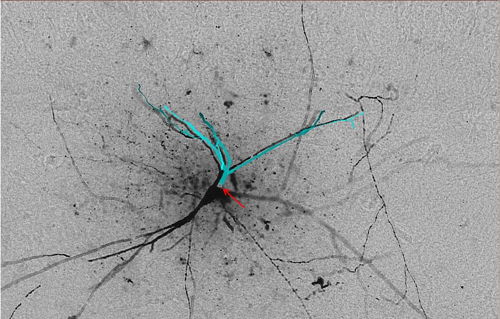
Gold standard reconstruction '01.swc' (light blue) from ‘Neuron 1’ is superimposed and offset (for visualization purposes) over image stack 'Section 01'. Arrow (red) points to the root of the reconstruction.
Neuron 1 Properties:
Individual Stack Information: All image stacks are 24 bits per pixel.
Section 1- Number of images: 110; Pixel Size (X,Y): (5360,3420)
Section 2- Number of images: 135; Pixel Size (X,Y): (5896,3040)
Section 3- Number of images: 120; Pixel Size (X,Y): (3752,2660)
Section 4- Number of images: 105; Pixel Size (X,Y): (4288,3040)
Section 5- Number of images: 82; Pixel Size (X,Y): (3752,2280)
Gold Standard Reconstruction Starting Coordinates (X and Y in pixels; Z in image sequence number, where top image Z = 0):
Section_1
S1_01 (X,Y,Z): (3475,634,66)
S1_02 (X,Y,Z): (3526,667,98)
S1_03 (X,Y,Z): (3625,747,105)
S1_04 (X,Y,Z): (3926,322,37)
S1_05 (X,Y,Z): (3504,706,32)
S1_06 (X,Y,Z): (2084,1229,110)
S1_07 (X,Y,Z): (3501,703,13)
S1_08 (X,Y,Z): (3412,613,12)
S1_09 (X,Y,Z): (3333,627,16)
S1_10 (X,Y,Z): (3433,695,58)
S1_11 (X,Y,Z): (2501,1700,109)
S1_12 (X,Y,Z): (2437,1376,58)
S1_13 (X,Y,Z): (2875,983,50)
S1_14 (X,Y,Z): (3339,631,103)
Section_2
S2_01 (X,Y,Z): (4183,563,80)
S2_02 (X,Y,Z): (4503,1172,113)
S2_03 (X,Y,Z): (4063,861,108)
S2_04 (X,Y,Z): (3907,869,110)
S2_05 (X,Y,Z): (3889,839,111)
S2_06 (X,Y,Z): (3613,902,112)
S2_07 (X,Y,Z): (3357,1573,116)
S2_08 (X,Y,Z): (1912,2550,0)
S2_09 (X,Y,Z): (2466,1512,115)
Section_3
S3_01 (X,Y,Z): (2341,286,117)
S3_02 (X,Y,Z): (2094,790,101)
S3_03 (X,Y,Z): (1001,1336,55)
S3_04 (X,Y,Z): (967,1807,119)
S3_05 (X,Y,Z): (760,1864,57)
S3_06 (X,Y,Z): (770,2001,60)
Section_4
S4_01 (X,Y,Z): (2478,702,23)
S4_02 (X,Y,Z): (2487,726,84)
S4_03 (X,Y,Z): (3024,336,16)
S4_04 (X,Y,Z): (2417,823,88)
S4_05 (X,Y,Z): (2465,723,16)
S4_06 (X,Y,Z): (2443,846,19)
S4_07 (X,Y,Z): (3373,1037,15)
S4_08 (X,Y,Z): (3513,1382,20)
S4_09 (X,Y,Z): (2045,547,5)
S4_10 (X,Y,Z): (1452,832,22)
S4_11 (X,Y,Z): (2938,1533,25)
S4_12 (X,Y,Z): (589,1871,26)
S4_13 (X,Y,Z): (866,2066,30)
S4_14 (X,Y,Z): (3262,1263,92)
Section_5
S5_01 (X,Y,Z): (2710,281,6)
S5_02 (X,Y,Z): (3534,1073,10)
S5_03 (X,Y,Z): (2725,1126,13)
S5_04 (X,Y,Z): (1885,703,5)
S5_05 (X,Y,Z): (1152,1174,12)
S5_06 (X,Y,Z): (336,1985,13)
Neuron 2 Properties:
Individual Stack Information: All image stacks are 24 bits per pixel.
Section 1- Number of images: 105; Pixel Size (X,Y): (4824,2660)
Section 2- Number of images: 110; Pixel Size (X,Y): (8040,2660)
Section 2 75% size- Number of images: 110 (X,Y); Pixel Size: (6030,1995)
Section 3- Number of images: 105; Pixel Size (X,Y): (4824,2660)
Section 4- Number of images: 99; Pixel Size (X,Y): (5360,3040)
Section 4 90% size- Number of images: 99; Pixel Size (X,Y): (4824,2736)
Gold Standard Reconstruction Starting Coordinates (X and Y in pixels; Z in image sequence number, where top image Z = 0):
Section_1
S1_01 (X,Y,Z): (1776,576,42)
S1_02 (X,Y,Z): (2059,675,71)
S1_03 (X,Y,Z): (1739,661,38)
S1_04 (X,Y,Z): (1466,753,75)
S1_05 (X,Y,Z): (1685,553,72)
S1_06 (X,Y,Z): (1472,1316,78)
S1_07 (X,Y,Z): (1441,1614,80)
S1_08 (X,Y,Z): (1350,1655,67)
Section_2
S2_01 (X,Y,Z): (6396,1060,109)
S2_02 (X,Y,Z): (5745,1086,42)
S2_03 (X,Y,Z): (4929,1379,109)
S2_04 (X,Y,Z): (4887,1967,48)
Section_2 (75% Scaled)
S2_01_75% (X,Y,Z): (4797,795,109)
S2_02_75% (X,Y,Z): (4309,815,42)
S2_03_75% (X,Y,Z): (3697,1034,109)
S2_04_75% (X,Y,Z): (3665,1475,48)
Section_3
S3_01 (X,Y,Z): (2004,783,24)
S3_02 (X,Y,Z): (420,1376,20)
S3_03 (X,Y,Z): (1525,812,93)
S3_04 (X,Y,Z): (1144,1213,30)
S3_05 (X,Y,Z): (2368,1266,26)
Section_4
S4_01 (X,Y,Z): (2801,1548,14)
S4_02 (X,Y,Z): (3283,1620,46)
S4_03 (X,Y,Z): (4101,336,80)
S4_04 (X,Y,Z): (2750,1595,78)
Section_4 (90% Scaled)
S4_01_90% (X,Y,Z): (2521,1394,14)
S4_02_90% (X,Y,Z): (2955,1451,46)
S4_03_90% (X,Y,Z): (3691,302,80)
S4_04_90% (X,Y,Z): (2475,1436,78)